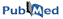|
|
The MC-Fold | MC-Sym pipeline is a web-hosted service for RNA secondary and tertiary structure
prediction. The pipeline means that the input sequence to MC-Fold outputs
secondary structures that are direct input to MC-Sym, which outputs tertiary structures.
The vertical bar '|' is the symbol indicating pipeline processing in Unix, which was invented by Malcolm D. McIlroy. Reference:
|
|
|
MC-Fold predicts secondary structures from sequence (black arrow).
Compared to classical secondary structures that include the AU, CG,
and Wobble GU base pairs that form the stems of the structure,
MC-Fold's secondary structures include the other base pairing patterns
as well. These are often called non-canonical base pairs.
⊕ MC-Fold's database folder Reference:
|
MC-Fold :: sequence -> [secondary-structure]Link to MC-Fold
♦ MC-Fold is available online via command line using the cURL program:
curl -Y 0 -y 300 -F "pass=lucy" -F "sequence=CCAGAUCUGAGCCUGGGAGCUCUCUGG" https://www.major.iric.ca/cgi-bin/MC-Fold/mcfold.static.cgi
♦ Compare the results of different secondary structure prediction programs using the following sequences:
- HIV-1 TAR RNA: CCAGAUCUGAGCCUGGGAGCUCUCUGG
- IRE with V-bulge: AUUAUCGGGAGCAGUGUCUUCCAUAAU
- IRE with W-bulge: AUUCUUGCUUCAACAGUGUUUGAACGGAA
- Rat 28S E-loop: GGGUGCUCAGUACGAGAGGAACCGCACCC
- C27A ScYLV pseudoknot: AGUGGCGCCGACCACUUAAAAACAACGG
♦ Alternative secondary structure prediction programs:
- ILM Ruan, Stormo, Zhang. NAR 2004.
- Sfold Ding, Lawrence. NAR 2003.
- Pfold Knudsen, Hein. NAR 2003.
- Mfold Zuker. NAR 2003.
- VSfold Dawson, Fujiwara, Kawai. PLoS One 2007.
- RNAfold Hofacker. NAR 2003.
- CONTRAfold Do, Woods, Batzoglou. Bioinformatics 2006.
- RNAstructure Mathews et al. PNAS 2004.
|
|
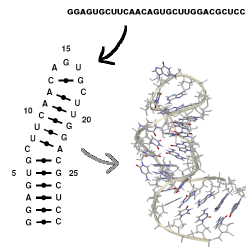
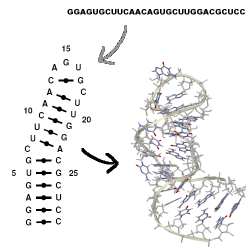
MC-Sym builds tertiary structures from MC-Fold's secondary structures, which are
provided as ASCII input scripts. No interactive computer graphics is needed.
MC-Sym explores exhaustively or probabilistically the conformational search
space of an RNA and produces structures that satisfies all input constraints.
⊕ MC-Sym's FAQ References:
|
MC-Sym :: secondary-structure -> [tertiary-structure]Link to MC-Sym
♦ Here is a list of MC-Sym input scripts and PDB references:
| PDB ID | MC-Sym | Reference | Type | Description | ||||
| 430D | 430D.mcc | 430D01.pdb | Hairpin | Rat 28S Loop E | ||||
| 1NBR | 1NBR.mcc | 1NBR01.pdb | Hairpin | Iron Responsive Element V-form | ||||
| 2HUA | 2HUA.mcc | 2HUA01.pdb | Hairpin | SCFV IRES Domain III | ||||
| 2GIO | 2GIO.mcc | 2GIO01.pdb | Hairpin | RNA Thermometer | ||||
| 2FDT | 2FDT.mcc | 2FDT01.pdb | Hairpin | UnaL2 Interspersed Element | ||||
| 2FEY | 2FEY.mcc | 2FEY01.pdb | Hairpin | Telomerase RNA Domain IV | ||||
| 2CD1 | 2CD1.mcc | 2CD101.pdb | Hairpin | RNase P RNA P4 | ||||
| 2EVY | 2EVY.mcc | 2EVY01.pdb | Hairpin | GNYA Tetraloop | ||||
| 2O33 | 2O33.mcc | 2O3301.pdb | Hairpin | U2 snRNA | ||||
| 2AHT | 2AHT.mcc | 2AHT01.pdb | Hairpin | Group II Intron Branchsite | ||||
| 1NYI | 1NYI.mcc | 1NYI01.pdb | Y-Shape | Hammerhead Ribozyme | ||||
| 2HGH | 2HGH.mcc | 2HGH01.pdb | Y-Shape | 5S rRNA | ||||
| 2AP5 | 2AP5.mcc | 2AP501.pdb | Pseudoknot | Yellow Leaf Virus | ||||
| 2P89 | [P1] [P1S] [P2] [P2S] | 2P8901.pdb | Complex | H/ACA Box | ||||
| [Whole] |
|
|
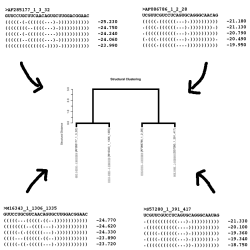
MC-Cons assigns a secondary structure for each sequence among the sequence's set of sub-optimal secondary structures, such that the set of assigned secondary structures maximizes the sum of pairwise structural similarity; the consensus assignment. MC-Cons can be seen as a filter between multiple-sequences and tertiary structure.
⊕ MC-Cons's user's guide Reference:
|
MC-Cons :: [(sequence, [secondary-structure])] -> [(sequence, secondary-structure)]Link to MC-Cons
♦ Alternative consensus structure and assignment programs:
- Marna (MC-Fold output friendly) Siebert, Backofen. Bioinformatics 2005.
- RNAshapes (sequence data only) Steffen et al. Bioinformatics 2006.
|
A tool for rendering dot-bracket structures is plot_rna of the CONTRAfold package (Do et al. Bioinformatics 2006).
|
|
Here is an input example:
>ce TrxR GACCUUUGGCUAAACUCCAUCGUGAGCGCCUCUGGUC ((((((((((..((((((...)))))))))))))))) -32.465 ((((((((((...(((((...).)))))))))))))) -32.570
♦ Alternative rendering programs with pseudoknot capability:
- PseudoViewer Byun, Han. NAR 2006. (online web server)
- jViz.Rna Wiese, Glen. IEEE CBSM 2006. (manual adjustments)
|
This program will generate an MC-Sym script that will build an RNA of the specified sequence and secondary structure. Please consider these points:
- Accepted symbols for sequence are: `A`, `C`, `G`, `U` and `T`.
- Accepted symbols for structure are: `.`, `(`, `[`, `)` and `]`.
- Isolated base pairs .(. .). are not modeled if they are not within NCMs.
- Structures with more than 40% in single-stranded (i.e. non-structures) are not modeled.
- Classic secondary structures are difficult to model properly.
- Pseudoknots are accepted, as long as the expression is properly balanced.
- For advanced modeling topics please visit our FAQ.
Here is an input example:
>ce TrxR GACCUUUGGCUAAACUCCAUCGUGAGCGCCUCUGGUC ((((((((((..((((((...))))))))))))))))
Here is another input example, with a pseudoknot:
>2TPK UGACCAGCUAUGAGGUCAUACAUCGUCAUAGC (((((.[[[[[[[))))).......]]]]]]]
|
This program will generate an MC-Sym script that will build an RNA double-helix.
♦ Alternative double-helix generators:
- PREDICTOR (DNA double-helix) Farwer et al. In Silico Biol 2007. (online web server)
|
This program will calculate the interaction network fidelity (INF) between two RNA 3-D structures based on their annotations.
|
This program will mutate the nucleobase identity to the specified sequence into the furnished 3D structure. This is useful to perform point mutations in a structure, or to replace non-canonical nucleobases by standard one.
Here is an input example:
Structure: PDB file 1EVV (without ions and water molecules). Sequence: GCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA
|
|
|
created: October 16, 2007
updated: August 05, 2009